library(stringr)
library(tidyverse)
library(data.table)
library(dbscan)
library(factoextra)
dt <- fread("input.txt", header = FALSE, colClasses = "character")
dt <- data.frame(str_split(dt$V1, "", simplify = TRUE))
dtPadTop <- dt[1,]
dtPadTop[!is.na(dtPadTop)] <- 10
dt <- rbind(rbind(dtPadTop, dt), dtPadTop)
dtPadSides <- dt[,1]
dtPadSides[!is.na(dtPadSides)] <- 10
dt <- cbind(cbind(dtPadSides, dt), dtPadSides)
dt <- data.frame(apply(dt, 2, as.numeric))Smoke Basin
Part 1
dt2 <- dt
coordX <- c()
coordY <- c()
rLow <- c()
for(r in 2:(nrow(dt2) - 1)) {
for(c in 2:(ncol(dt2) - 1)) {
if(dt2[r,c] == min(dt[(r-1):(r+1),(c-1):(c+1)])) {
rLow <- append(rLow, dt2[r,c])
coordX <- append(coordX, c)
coordY <- append(coordY, r)
}
}
}
s <- 0
for(n in 1:length(rLow)) {
s <- s + (rLow[n] + 1)
}
s[1] 575Part 2
dt2 <- dt
dt2[dt2 == 9 | dt2 == 10] <- NA
coordX <- c()
coordY <- c()
rLow <- c()
for(r in 2:(nrow(dt2) - 1)) {
for(c in 2:(ncol(dt2) - 1)) {
if(!is.na(dt2[r,c])) {
rLow <- append(rLow, dt2[r,c])
coordX <- append(coordX, c)
coordY <- append(coordY, r)
}
}
}
dt3 <- data.frame("x" = coordX, "y" = coordY, "d" = rLow)
dt3 <- dt3[dt3$d <= 8,]
db <- dbscan(dt3[,1:2], eps = 1, minPts = 0)
dt3$cluster <- db$cluster
dt <- dt3 %>%
group_by(cluster) %>%
count() %>%
filter(cluster != 0) %>%
arrange(desc(n)) %>%
head(3)
prod(dt$n)[1] 1019700dt3$cluster <- ifelse(dt3$cluster %in% dt$cluster, dt3$cluster, 0)
dt3 %>%
mutate(cluster = as.factor(cluster)) %>%
ggplot() +
geom_point(aes(x = x, y = y, color = cluster))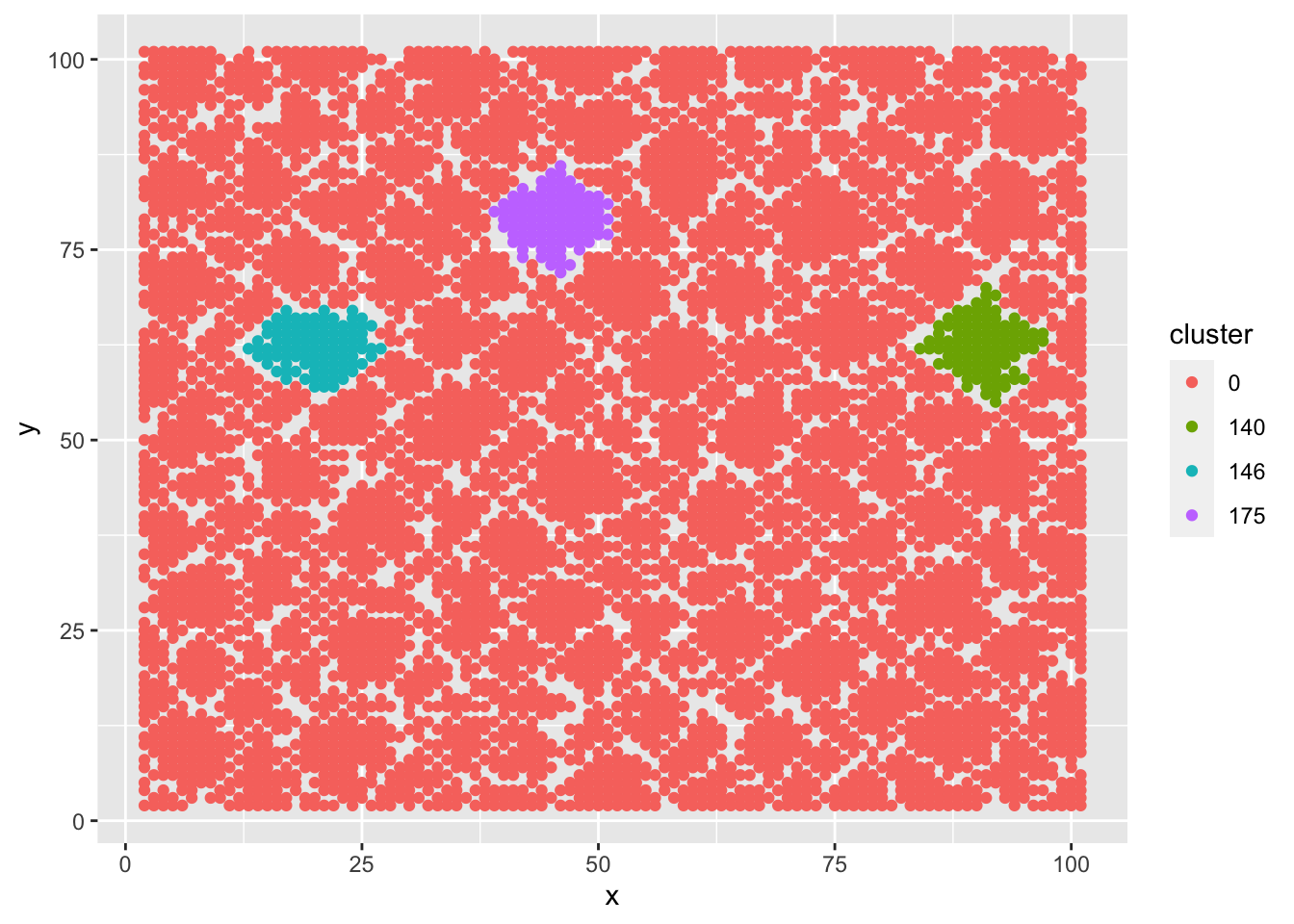
Saving 7 x 5 in image